Previous Research
Background
Small non-coding RNAs have been discovered almost two decades ago. They play crucial roles in regulating development and differentiation, cellular defense against viruses and in maintaining genomic integrity. At the core of small RNA pathways lies an effector comples consisting of the small RNA, which conveys sequence specificity and the associated Argonatue protein, which is responsible for the effector function. The best studied small RNAs are microRNAs, which associate with the Ago clade of Argonaute proteins and regulate gene expression of host genes through sequence specific interaction with mRNAs leading to translational inhibition and target destabilization. Small interfering (si)RNAs are similar in their biogenesis and protein partners to miRNAs and their main function is believed to be targeting of foreign mRNAs followed by endonucleolytic cleavage and destruction of target mRNA. siRNAs have also been extensively used in biomedical research for sequence-specific knockdown of genes of interest in a process called RNA interference (RNAi). In S. pombe and plants siRNAs are known to also act in the nucleus to induce heterochromatinization of targets thereby leading to transcriptional silencing.
Recently, a new class of small RNAs, called Piwi-interacting or piRNAs, has been described. As their name implies, they associate with the Piwi clade of Argonaute proteins. Piwi proteins and piRNAs are restricted to the germline of metazoans and they function in a conserved pathway that is crucial for repressing mobile genomic elements. Uncontrolled transposon activity leads to mutations and genome instability. This is especially harmful in germline cells because germline mutations are transmitted to offspring. Mutations in piwi genes lead to failure of gametogenesis and sterility in both Drosophila and mouse. In the cytoplasm piwi proteins mediate cleavage of transposon transcripts, which are recognized through sequence complementarity to the piRNAs. Piwi proteins also function in the nucleus. In both Drosophila and mammals one member of the piwi clade is nuclear. The mouse piRNA pathway was shown to be required for de novo DNA methylation and silencing of TEs. The exact mechanism of this process is, however, not known.
Our recent studies have revealed that:
Piwi induces transcriptional silencing and chromatin changes
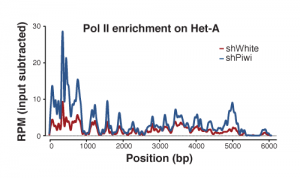
Piwi induces transcriptional silencing and establishment of repressive chromatin over TE targets: Using RNAi, we tested the effect of Piwi on transcription and chromatin state of the targets, transposable elements. We found that based on Pol II ChIP-seqtranscription rate is repressed by Piwi and that this repression correlates with the establishment of a repressive chromatin on targets.
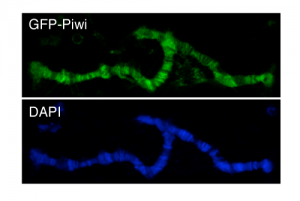
Piwi-associated piRNAs are required for recruiting Piwi to genomic targets: Based on the correspondence of targets transcriptionally silenced by Piwi and the Piwi-associated piRNAs we hypothesized that piRNAs are required for targeting Piwi to the genomic sites. We used reporter-based assays to confirm the role of piRNAs in recruiting Piwi to its genomic targets. We have also used this reporter system to identify additional chromatin changes that resulted from Piwi recruitment such as changes in some chromatin marks and recruitment of additional factors such as Rhino and Cuff. 
The piRNA pathway represents a true transgenerational epigenetic phenomenon
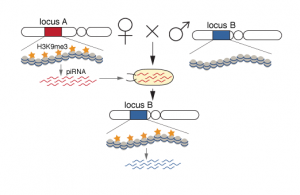 Certain RNAs are transmitted from one generation to the next and can provide an ideal means to transmit epigenetic memory. Indeed, in Drosophila piRNA-Piwi complexes are deposited into the oocytes and are present in the early embryo. These piRNA-Piwi complexes accumulate in the pole plasm, which gives rise to the gonads. Inherited piRNAs carry epigenetic information that determines target silencing in the progeny, but the mechanism by which the piRNAs impact targets was not known. In close collaboration, the Aravin lab and we have shown that inherited piRNAs affect the chromatin of target loci. We used D. melanogaster strains, which contain an identical transgene inserted into two different genomic loci. Germ cells generate abundant piRNAs from the transgene in one locus, while not from the other locus. Importantly, the inactive locus can be converted into an “active” locus by exposure to maternally inherited cytoplasm, carrying piRNAs homologous to the locus (deposited by the mother carrying the active locus). We found that the production of piRNAs from the active locus and conversion of the inactive locus to an active one (mediated by trans-generational piRNAs), was accompanied by an increase in H3K9me3 on the transgene. Therefore, exposure of an inactive locus to homologous piRNAs inherited from the previous generation leads to installment of H3K9me3 and conversion of the locus to an active piRNA-producing locus.
Certain RNAs are transmitted from one generation to the next and can provide an ideal means to transmit epigenetic memory. Indeed, in Drosophila piRNA-Piwi complexes are deposited into the oocytes and are present in the early embryo. These piRNA-Piwi complexes accumulate in the pole plasm, which gives rise to the gonads. Inherited piRNAs carry epigenetic information that determines target silencing in the progeny, but the mechanism by which the piRNAs impact targets was not known. In close collaboration, the Aravin lab and we have shown that inherited piRNAs affect the chromatin of target loci. We used D. melanogaster strains, which contain an identical transgene inserted into two different genomic loci. Germ cells generate abundant piRNAs from the transgene in one locus, while not from the other locus. Importantly, the inactive locus can be converted into an “active” locus by exposure to maternally inherited cytoplasm, carrying piRNAs homologous to the locus (deposited by the mother carrying the active locus). We found that the production of piRNAs from the active locus and conversion of the inactive locus to an active one (mediated by trans-generational piRNAs), was accompanied by an increase in H3K9me3 on the transgene. Therefore, exposure of an inactive locus to homologous piRNAs inherited from the previous generation leads to installment of H3K9me3 and conversion of the locus to an active piRNA-producing locus.