Our Approach
Biochemistry
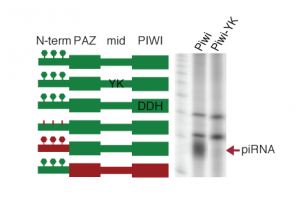
We apply biochemical approaches to identify protein-protein and protein-RNA interactions. Our methods focus on identifying transient protein-protein interaction partners in the piRNA pathway by utilizing crosslinking strategies during the immunoprecipitations followed by mass spectrometry. We complement these experiments with fluorescent microscopy based techniques that can detect weak or transient interactions. For RNA-protein interactions we apply the complementary strategies of identifying RNAs bound to specific factors (RIP, CLIP coupled with high throughput sequencing) and proteins associated with specific RNA (RAP coupled with mass spec). We are also functionally dissecting proteins of the piRNA pathway by testing the role of different protein domains and amino acid residues (and their posttranslational modifications) in protein function.
Molecular and Cell Biology
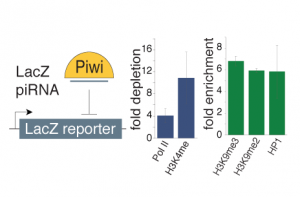
We are trying to understand how the piRNA pathway regulates expression of its targets. We are testing the impact of the nuclear Piwi protein on the chromatin landscape of its genomic targets and are identifying novel factors of the pathway using ChIP-based approaches. We are also analyzing how the chromatin environment impacts the emerging RNA using different RNA library cloning strategies and characterizing biochemical and functional properties of RNAs. We combine these experiments with genetic tools to identify factors that shape our transcriptome.
Fluorescent Imaging
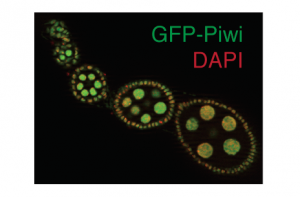
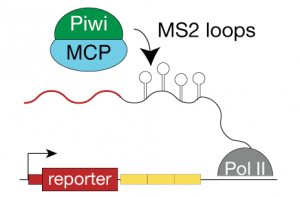
We use in vivo fluorescent microscopic techniques as well as immunofluorescence in flies and cell culture to complement our biochemical approaches. We have generated a wide range of transgenic fly lines with fluorescently tagged piRNA pathway components to assess their localization, expression level and dynamics under physiological conditions and upon perturbing the pathway. We also use fluorescent reporter systems to identify transient interactions of factors within the pathway.
Genetics
We use fly as a model system to dissect small RNA pathways. We apply both forward and reverse genetics to identify factors involved in the piRNA pathway and to dissect their role. For functional assays we use RNAi and more recently targeted mutagenesis using the CRISPR technique. We also use genetic tools to generate tagged transgenes both for biochemical and microscopic dissection of the piRNA pathway and more generally of RNA biology.
Genomics / Transcriptomics
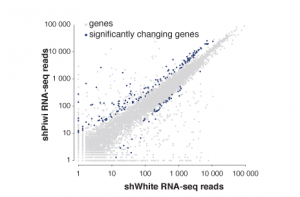
The readout of many of our experiments is high throughput sequencing data mostly from ChIP-seq and diverse RNA-seq methods. We utilize systems biology to integrate diverse datasets to comprehensively characterize pathways acting and interacting in regulating gene expression on multiple levels. In collaboration with other labs at Caltech we are developing computational tools to analyze, compare and integrate these diverse datasets.
